Introduction to R¶
Loading libraries¶
Some R functions are in libraries that have to be explicitly loaded. You
do this with the library command. The next cell shows an example of
loading the genefilter library which contains the rowttests
fucntion that we will be using later.
library(genefilter)
Attaching package: ‘genefilter’
The following object is masked from ‘package:base’:
anyNA
Installing libraries¶
Most libraries have been pre-installed on the VM. However, if you need
to install a new library, all you need is to use the
install.packages() function. For example, you can install the MASS
(Modern Applied Statistics in S) library like so:
install.packages("MASS", repos = "http://cran.r-project.org")
Some specialized libraries come from the R BioConductor project, and the installation processs is slgihtly different. This will be covered in a later session.
Printing results¶
By default, the result on the last line is displayed in the Jupyter
notebook. To see other lines, use the print function.
print(1 * 2 * 3 *4)
print(11 %% 3 == 1)
10 > 1
[1] 24
[1] FALSE
Getting help¶
Type soemthing and hit the tab key - this will either complete the R command for you (if unique) or present a list of opitons.
Use ?topic to get a description of topic, or more verbosely, you can
also use help(topic).
?seq
| seq {base} | R Documentation |
Sequence Generation
Description
Generate regular sequences. seq is a standard generic with a
default method. seq.int is a primitive which can be
much faster but has a few restrictions. seq_along and
seq_len are very fast primitives for two common cases.
Usage
seq(...)
## Default S3 method:
seq(from = 1, to = 1, by = ((to - from)/(length.out - 1)),
length.out = NULL, along.with = NULL, ...)
seq.int(from, to, by, length.out, along.with, ...)
seq_along(along.with)
seq_len(length.out)
Arguments
... |
arguments passed to or from methods. |
from, to |
the starting and (maximal) end values of the
sequence. Of length |
by |
number: increment of the sequence. |
length.out |
desired length of the sequence. A
non-negative number, which for |
along.with |
take the length from the length of this argument. |
Details
Numerical inputs should all be finite (that is, not infinite,
NaN or NA).
The interpretation of the unnamed arguments of seq and
seq.int is not standard, and it is recommended always to
name the arguments when programming.
seq is generic, and only the default method is described here.
Note that it dispatches on the class of the first argument
irrespective of argument names. This can have unintended consequences
if it is called with just one argument intending this to be taken as
along.with: it is much better to use seg_along in that
case.
seq.int is an internal generic which dispatches on
methods for "seq" based on the class of the first supplied
argument (before argument matching).
Typical usages are
seq(from, to) seq(from, to, by= ) seq(from, to, length.out= ) seq(along.with= ) seq(from) seq(length.out= )
The first form generates the sequence from, from+/-1, ..., to
(identical to from:to).
The second form generates from, from+by, ..., up to the
sequence value less than or equal to to. Specifying to -
from and by of opposite signs is an error. Note that the
computed final value can go just beyond to to allow for
rounding error, but is truncated to to. (‘Just beyond’
is by up to 1e-10 times abs(from - to).)
The third generates a sequence of length.out equally spaced
values from from to to. (length.out is usually
abbreviated to length or len, and seq_len is much
faster.)
The fourth form generates the integer sequence 1, 2, ...,
length(along.with). (along.with is usually abbreviated to
along, and seq_along is much faster.)
The fifth form generates the sequence 1, 2, ..., length(from)
(as if argument along.with had been specified), unless
the argument is numeric of length 1 when it is interpreted as
1:from (even for seq(0) for compatibility with S).
Using either seq_along or seq_len is much preferred
(unless strict S compatibility is essential).
The final form generates the integer sequence 1, 2, ...,
length.out unless length.out = 0, when it generates
integer(0).
Very small sequences (with from - to of the order of 10^{-14}
times the larger of the ends) will return from.
For seq (only), up to two of from, to and
by can be supplied as complex values provided length.out
or along.with is specified. More generally, the default method
of seq will handle classed objects with methods for
the Math, Ops and Summary group generics.
seq.int, seq_along and seq_len are
primitive.
Value
seq.int and the default method of seq for numeric
arguments return a vector of type "integer" or "double":
programmers should not rely on which.
seq_along and seq_len return an integer vector, unless
it is a long vector when it will be double.
References
Becker, R. A., Chambers, J. M. and Wilks, A. R. (1988) The New S Language. Wadsworth & Brooks/Cole.
See Also
The methods seq.Date and seq.POSIXt.
:,
rep,
sequence,
row,
col.
Examples
seq(0, 1, length.out = 11) seq(stats::rnorm(20)) # effectively 'along' seq(1, 9, by = 2) # matches 'end' seq(1, 9, by = pi) # stays below 'end' seq(1, 6, by = 3) seq(1.575, 5.125, by = 0.05) seq(17) # same as 1:17, or even better seq_len(17)
Use example to see usage examples.
example(seq)
seq> seq(0, 1, length.out = 11)
[1] 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0
seq> seq(stats::rnorm(20)) # effectively 'along'
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20
seq> seq(1, 9, by = 2) # matches 'end'
[1] 1 3 5 7 9
seq> seq(1, 9, by = pi) # stays below 'end'
[1] 1.000000 4.141593 7.283185
seq> seq(1, 6, by = 3)
[1] 1 4
seq> seq(1.575, 5.125, by = 0.05)
[1] 1.575 1.625 1.675 1.725 1.775 1.825 1.875 1.925 1.975 2.025 2.075 2.125
[13] 2.175 2.225 2.275 2.325 2.375 2.425 2.475 2.525 2.575 2.625 2.675 2.725
[25] 2.775 2.825 2.875 2.925 2.975 3.025 3.075 3.125 3.175 3.225 3.275 3.325
[37] 3.375 3.425 3.475 3.525 3.575 3.625 3.675 3.725 3.775 3.825 3.875 3.925
[49] 3.975 4.025 4.075 4.125 4.175 4.225 4.275 4.325 4.375 4.425 4.475 4.525
[61] 4.575 4.625 4.675 4.725 4.775 4.825 4.875 4.925 4.975 5.025 5.075 5.125
seq> seq(17) # same as 1:17, or even better seq_len(17)
[1] 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17
To search for all functions with the phrase topic, use
apropos(topic).
apropos("seq")
- 'seq'
- 'seq_along'
- 'seq.Date'
- 'seq.default'
- 'seq.int'
- 'seq_len'
- 'seq.POSIXt'
- 'sequence'
For a broader search using “fuzzy” matching, use ?? prepended to the
search term.
??seq
Finally, you can always use a search engine - e.g. Googling for “how to generate random numbers from a normal distribution in R” returens:
R Learning Module: Probabilities and Distributions www.ats.ucla.edu › stat › r › modules University of California, Los Angeles Generating random samples from a normal distribution ... (For more information on the random number generator used in R please refer to the help pages for the ...
R: The Normal Distribution https://stat.ethz.ch/R-manual/R-devel/library/.../Normal.html ETH Zurich Normal {stats}, R Documentation ... quantile function and random generation for the normal distribution with mean equal to mean and ... number of observations.
CASt R: Random numbers astrostatistics.psu.edu/.../R/Random.html Pennsylvania State University It is often necessary to simulate random numbers in R. There are many functions available to ... Let’s consider the well-known normal distribution as an example: ?Normal .... Creating functions in R, as illustrated above, is a common procedure.
Work!¶
These ntoebooks will be interspersed with many such Work! sections,
since the only way to learn R is by using R. Remember to use TAB and
? and apropos whenever you’re stuck. If you don’t like to work
so much, reanme them as Play! sections since in our world
work == play.
If you put USD 100 in a bank that pays5 percent interest compounded daily, how much will you have in the bank at the end of one year? Recall from high school math that
where \(A\) is the amount, \(P\) is the principal, \(r\) is the interest rate, \(n\) is the compoundings per period and \(m\) is the number of periods.
What is the (Euclideean) distance between two points at (1,5) and (4,8)? Recall that the Eucliean distacne between two points (x1, y1) and (x2, y2) is given by the Pythagorean theorem:
Creating and using sequences¶
1:4
- 1
- 2
- 3
- 4
seq(2, 12, 3)
- 2
- 5
- 8
- 11
seq(from=0, to=pi, by=pi/4)
- 0
- 0.785398163397448
- 1.5707963267949
- 2.35619449019234
- 3.14159265358979
seq(from=0, to=pi, length.out = 5)
- 0
- 0.785398163397448
- 1.5707963267949
- 2.35619449019234
- 3.14159265358979
Vectorized operations¶
x <- 1:10
y <- 1:10
x * y
- 1
- 4
- 9
- 16
- 25
- 36
- 49
- 64
- 81
- 100
x <- seq(0, 2*pi, length.out = 5)
sin(x)
- 0
- 1
- 1.22464679914735e-16
- -1
- -2.44929359829471e-16
Functions¶
A function is code tha behaves like a black box - you give it some input
and it returns some output. Functions are essenital because they allow
you to design use potentially complicated algorithms without having to
keep track of the algorithmic details - all you need to remeber are the
function name (which you can search with apropos or prepnding ??
to the search term), the input arguments (TAB provides hings), and what
type of output will be generated. Most of R’s usefulness comes from the
large collection of functions that it provides - for example,
library(), seq(), as.fractions() are all functions. You can
also write your own R functions - this will be covered in detail in a
later session - but we show the basics here so that you can follow the
statistics lecutures.
A function has the following structure
function_name <- function(arguments) {
body_of_function
return(result)
}
The simple example below will make this explicit.
my.func <- function(a, b) {
c <- a * b
return(a + b - c)
}
my.func(5, 4)
my.func(b=4, a=5)
Work!¶
What is the sum of all the numbers from 1 to 100?
What is the sum of the natural log of all the numbers from 1 to 100?
What is the sum of all the odd numbers from 1 to 10?
Write a function that returns the sum of all the odd numbers from a
to b, where a and b are the minimum and maximum values of a
sequence of integers. For example, if the funciton was called
sum.odd, then you would answer the previous question by
sum.odd(1, 100)
Graphics¶
x <- seq(0, 2*pi, length.out = 100)
plot(x, sin(x), main="Graph of Sin(x)")
par(mfrow=c(2,2))
plot(x, sin(x), main="Graph of Sin(x)")
plot(x, cos(x), main="Graph of Cos(x)", type="o", col="blue", lty=2)
plot(x, tan(x), main="Graph of Tan(x)", type="l", col="red")
The concept of “saving” a graphical plot in R involves the following steps
- open a “device” to plot to (the next shows a
pdfdevice)- this includes the filename and other configuration parameters such as width and height
- write all the plotting and customization commands as usual
- close the device with
dev.close()- this writes the plot to file
pdf("trig.pdf", width=12, height=4)
par(mfrow=c(1,3))
plot(x, sin(x), main="Graph of Sin(x)")
plot(x, cos(x), main="Graph of Cos(x)", type="o", col="blue", lty=2)
plot(x, tan(x), main="Graph of Tan(x)", type="l", col="red")
dev.off()
?device
| Devices {grDevices} | R Documentation |
List of Graphical Devices
Description
The following graphics devices are currently available:
-
pdfWrite PDF graphics commands to a file -
postscriptWrites PostScript graphics commands to a file -
xfigDevice for XFIG graphics file format -
bitmapbitmap pseudo-device viaGhostscript(if available). -
pictexWrites TeX/PicTeX graphics commands to a file (of historical interest only)
The following devices will be functional if R was compiled to use them (they exist but will return with a warning on other systems):
-
X11The graphics device for the X11 windowing system -
cairo_pdf,cairo_psPDF and PostScript devices based on cairo graphics. -
svgSVG device based on cairo graphics. -
pngPNG bitmap device -
jpegJPEG bitmap device -
bmpBMP bitmap device -
tiffTIFF bitmap device -
quartzThe graphics device for the OS X native Quartz 2d graphics system. (This is only functional on OS X where it can be used from theR.appGUI and from the command line: but it will display on the local screen even for a remote session.)
Details
If no device is open, using a high-level graphics function will cause
a device to be opened. Which device is given by
options("device") which is initially set as the most
appropriate for each platform: a screen device for most interactive use and
pdf (or the setting of R_DEFAULT_DEVICE)
otherwise. The exception is interactive use under Unix if no screen
device is known to be available, when pdf() is used.
It is possible for an R package to provide further graphics devices and several packages on CRAN do so. These include other devices outputting SVG and PGF/TiKZ (TeX-based graphics, see http://pgf.sourceforge.net/).
See Also
The individual help files for further information on any of the
devices listed here;
X11.options, quartz.options,
ps.options and pdf.options for how to
customize devices.
dev.interactive,
dev.cur, dev.print,
graphics.off, image,
dev2bitmap.
capabilities to see if X11,
jpeg png, tiff,
quartz and the cairo-based devices are available.
Examples
## Not run: ## open the default screen device on this platform if no device is ## open if(dev.cur() == 1) dev.new() ## End(Not run)
Work!¶
Generate 100 random numbers from a standard normal distribution and
assign to a variable r. Plot a histogram of the distribuiton of
numbers in r.
Save the histogram you just geneerated as a PNG file.
Plot the sin, cos and tan functions from the examples for x
beteen 0 and 2\(\pi\), but place them all in the same plot using
different colors and line styles to distinguish them. Add an appropriate
figure legend.
Introudction to simulations¶
The statistics lectures will use statistical simulations extensively to illustrate key concepts. Basically, a simulaition is the use of random numbers drawn from some probability distribution to simulate experimental results. Simulations are a wonderful way to learn more about probability distributions and the concepts behind statistical inference.
set.seed(123) # fix random number seed for reproducibility
(x <- c("H", "T"))
- 'H'
- 'T'
sample(x, 20, replace=TRUE)
- 'T'
- 'H'
- 'H'
- 'T'
- 'T'
- 'H'
- 'T'
- 'T'
- 'H'
- 'T'
- 'T'
- 'T'
- 'H'
- 'H'
- 'H'
- 'T'
- 'T'
- 'H'
- 'T'
- 'H'
(x <- 1:6)
- 1
- 2
- 3
- 4
- 5
- 6
sample(x, 10, replace=FALSE)
Error in sample.int(length(x), size, replace, prob): cannot take a sample larger than the population when 'replace = FALSE'
sample(x)
- 1
- 6
- 4
- 3
- 2
- 5
sample(x)
- 3
- 4
- 2
- 1
- 6
- 5
sample(x, size = 3)
- 3
- 2
- 5
sample(x, size=10, replace=TRUE, prob=c(10,10,10,1,1,1))
- 3
- 3
- 4
- 1
- 2
- 1
- 1
- 3
- 1
- 3
Continuous distributions¶
par(mfrow=c(2,2)) # arrange plots in a 2 x 2 grid
x <- rnorm(n=100, mean=0, sd=1)
hist(x, main="Normal distribution")
rug(x)
x <- rexp(n=100, rate = 1)
hist(x, main="Exponenital distribution")
rug(x)
x <- runif(n=100, min=0, max=10)
hist(x, main="Uniform distribution")
rug(x)
x <- rbeta(n=100, shape1 = 0.5, shape2=0.5)
hist(x, main="Beta distribution")
rug(x)
Discrete distributions¶
par(mfrow=c(2,2)) # arrange plots in a 2 x 2 grid
x <- rpois(n=100, lambda = 1)
barplot(table(x), main="Poisson distribution")
x <- rbinom(n=100, size=10, prob=0.3)
barplot(table(x), main="Bionmial distribution")
x <- rnbinom(n=100, size=10, prob=0.3)
barplot(table(x), main="Negative binomial distribution")
x <- rgeom(n=100, prob=0.3)
barplot(table(x), main="Geometric distribution")
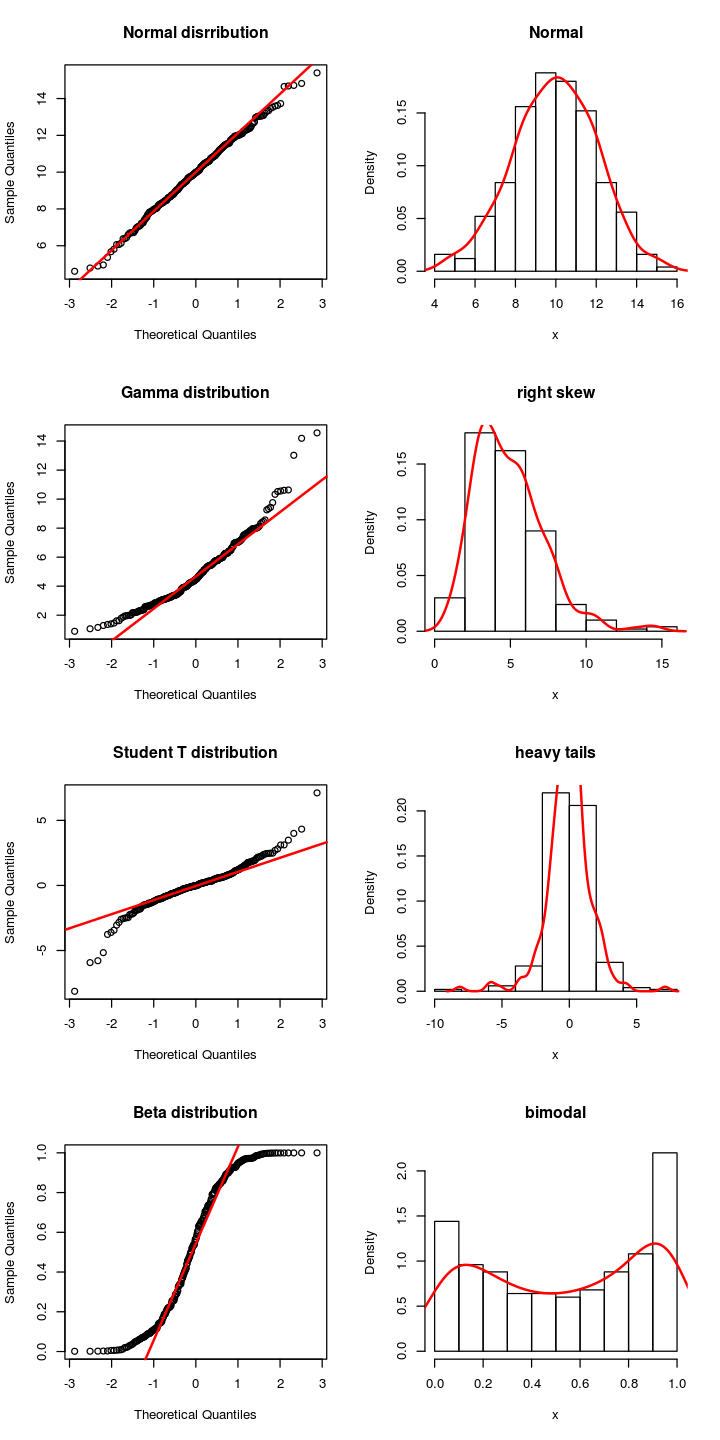
Checking distributions with qqplot¶
Most commonly, we want to check if data comes from a normal
distribution. To do this, we can plot the sample quantiles against the
theoretical quantiles - if the data is normally distributed, this should
not deviate far from a straight line. To chack against a normal
distribution, use qqnorm(x) where x is the sample to be tested. More
generally, you can test if x comes from any distribution using
qqplot(x, dist).
options(repr.plot.width = 6)
options(repr.plot.height = 12)
par(mfrow=c(4,2))
n <- 250
x <- rnorm(n=n, mean=10, sd=2)
qqnorm(x, main="Normal disrribution")
qqline(x, col="red", lwd=2)
hist(x, probability = TRUE, main="Normal")
lines(density(x), col="red", lwd=2)
x <- rgamma(n=n, scale=1, shape=5)
qqnorm(x, main="Gamma distribution")
qqline(x, col="red", lwd=2)
hist(x, probability = TRUE, main="right skew")
lines(density(x), col="red", lwd=2)
x <- rt(n=n, df=3)
qqnorm(x, main="Student T distribution")
qqline(x, col="red", lwd=2)
hist(x, probability = TRUE, main="heavy tails")
lines(density(x), col="red", lwd=2)
x <- rbeta(n=n, shape1 = 0.5, shape2=0.5)
qqnorm(x, main="Beta distribution")
qqline(x, col="red", lwd=2)
hist(x, probability = TRUE, main="bimodal")
lines(density(x), col="red", lwd=2)
A simple simulation¶
How frequently do we find significant differences (p < 0.05) between two groups when they are drawn from the same distribuiton if we use a t test?
As a bonus, this code snippet also shows a simple way to time chunks of code.
Digression - using Loops¶
To repeat an action many times, we use loops. The most useful loop construct in R is the for loop. A simple example shows how to use it.
for (i in 1:4) {
print(c(i, i*i))
}
[1] 1 1
[1] 2 4
[1] 3 9
[1] 4 16
Loops many include an if condition to only run the code if some condition is met.
for (i in 1:4) {
if (i %% 2 == 0) { # is i an even number?
print(c(i, i*i))
}
}
[1] 2 4
[1] 4 16
Now that we know how to use loops, let’s write the actural simulation.
nexpts = 10000
nsamps = 100
s = 0
alpha = 0.05
ptm <- proc.time()
for (i in 1:nexpts) {
u <- rnorm(nsamps)
v <- rnorm(nsamps)
if (t.test(u, v)$p.value < alpha)
s = s + 1
}
print(s/nexpts)
proc.time() - ptm
[1] 0.047
user system elapsed
2.544 0.006 2.551
Looping in R can be slow - here is a fast vectorized version¶
ptm <- proc.time()
x <- matrix(rnorm(2*nexpts*nsamps), nexpts, 2*nsamps)
grp <- factor(rep(0:1, c(nsamps, nsamps)))
print(sum(rowttests(x, grp)$p.value < alpha)/nexpts)
proc.time() - ptm
[1] 0.0507
user system elapsed
0.544 0.017 0.562
Work!¶
How frequently do we find significant differences (p < 0.05) between two groups when group 1 is drawn form a \(\mathcal{N}(0, 1)\) distribution and group 2 from an \(\mathcal{N}(0.2, 1)\) distribution? Write one version using explicit looping and another using vecgorized code, using 10000 experiments and 100 samples per group.