Base Graphics¶
Building Graphs¶
Incremental building of features into graphs.
References
- Graphical parameters
- Colors
- Named colors - type
colors() - Using color scales and plattes in R
?plot?par- Specific options e.g.
?pch
Basic plots¶
head(mtcars)
| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mazda RX4 | 21 | 6 | 160 | 110 | 3.9 | 2.62 | 16.46 | 0 | 1 | 4 | 4 |
| Mazda RX4 Wag | 21 | 6 | 160 | 110 | 3.9 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| Datsun 710 | 22.8 | 4 | 108 | 93 | 3.85 | 2.32 | 18.61 | 1 | 1 | 4 | 1 |
| Hornet 4 Drive | 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| Hornet Sportabout | 18.7 | 8 | 360 | 175 | 3.15 | 3.44 | 17.02 | 0 | 0 | 3 | 2 |
| Valiant | 18.1 | 6 | 225 | 105 | 2.76 | 3.46 | 20.22 | 1 | 0 | 3 | 1 |
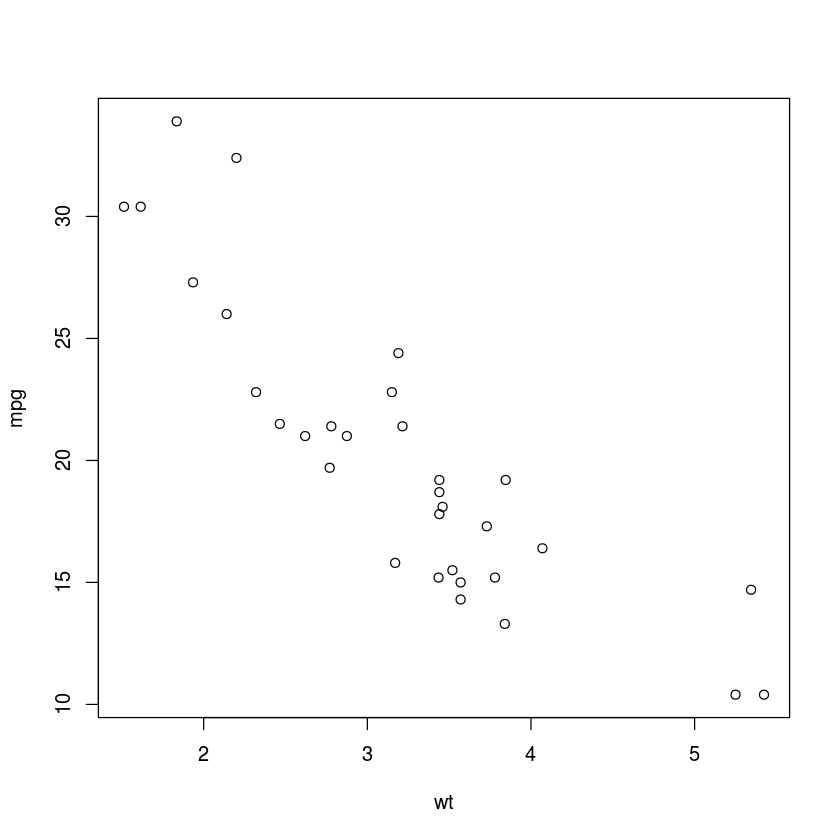
with(mtcars, plot(wt, mpg))
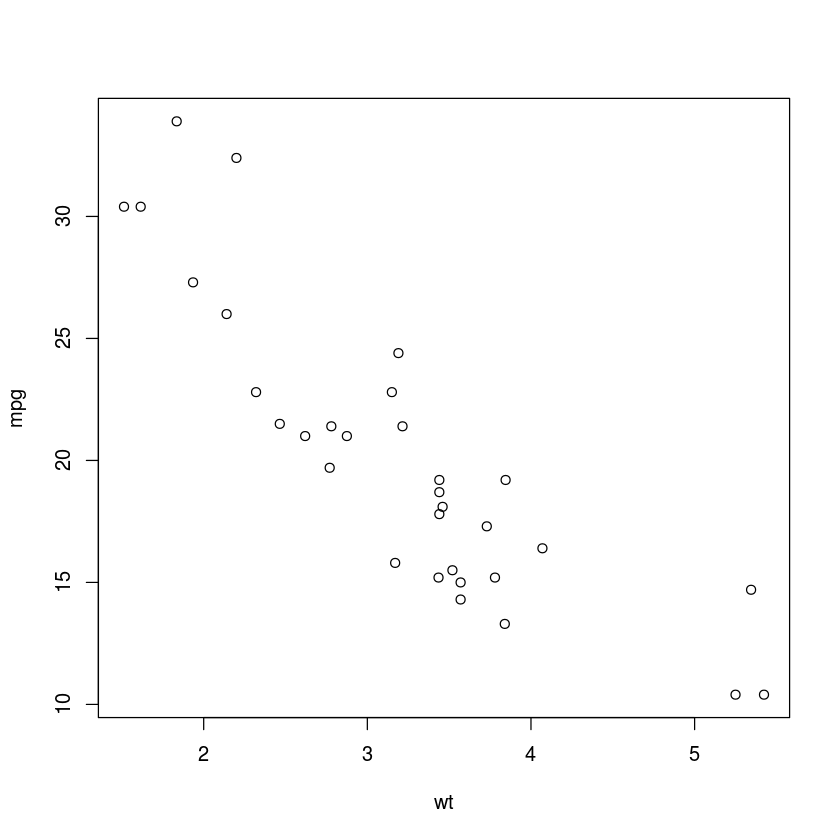
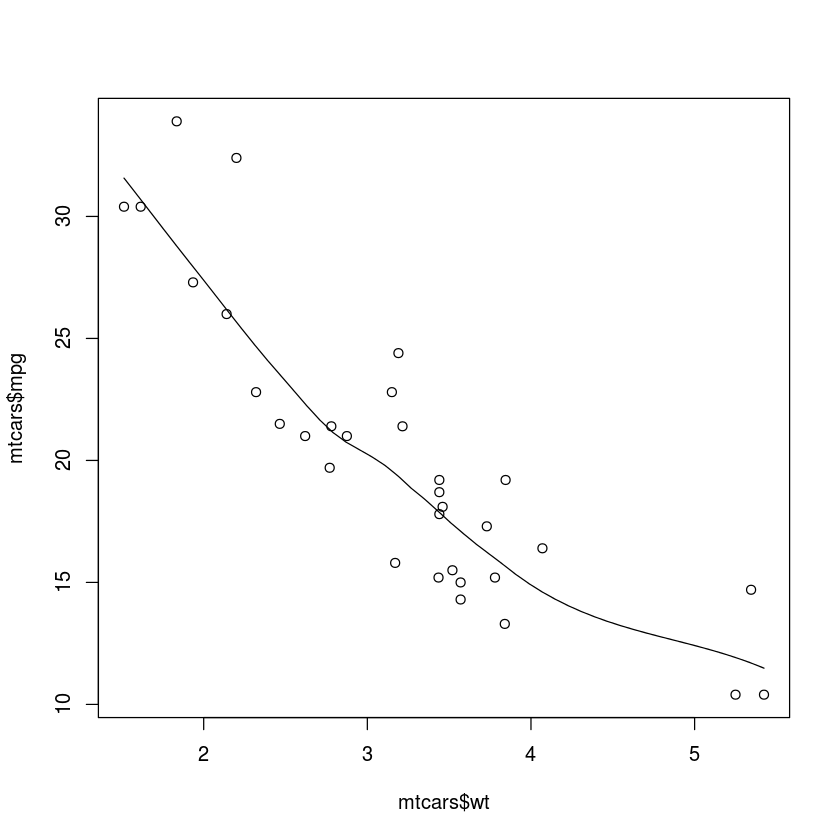
scatter.smooth(mtcars$wt, mtcars$mpg)
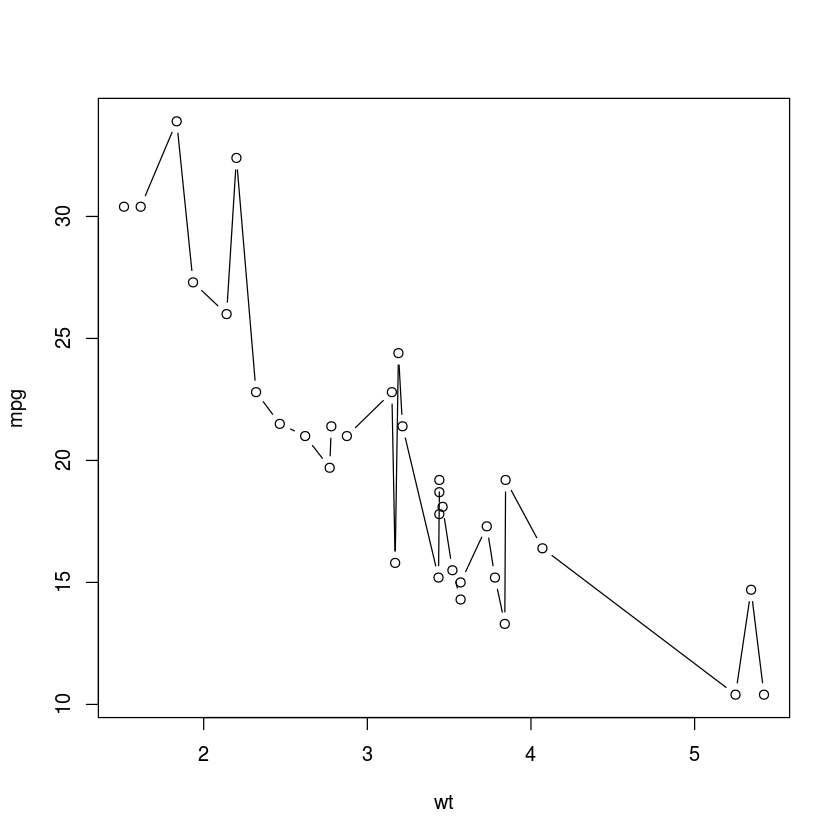
df <- mtcars[order(mtcars$wt),]
with(df, plot(wt, mpg, type="b"))
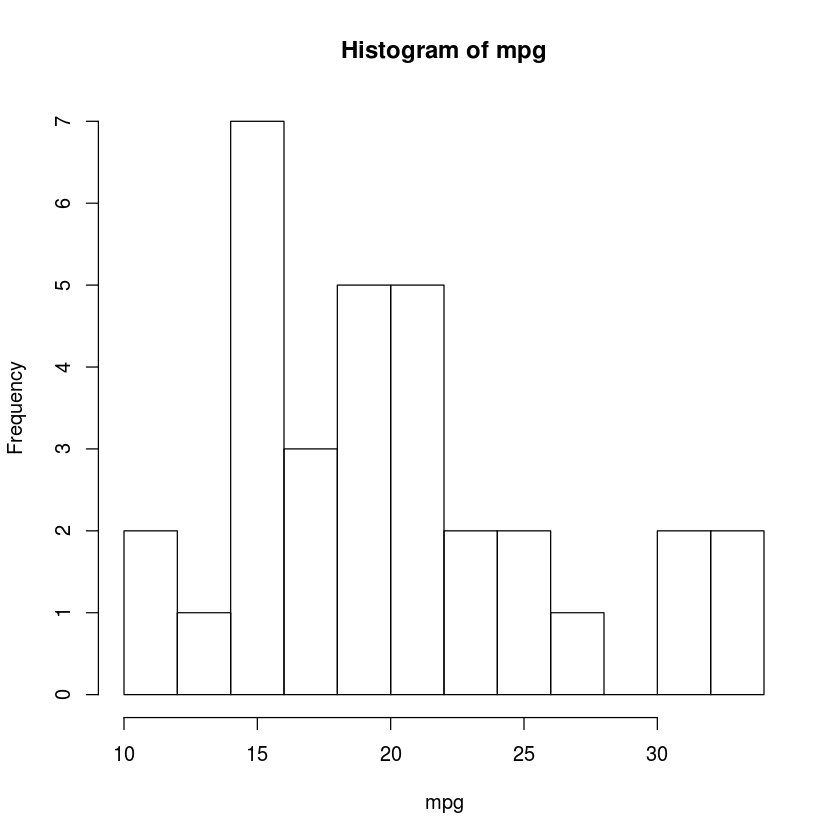
with(mtcars, hist(mpg, breaks=10))
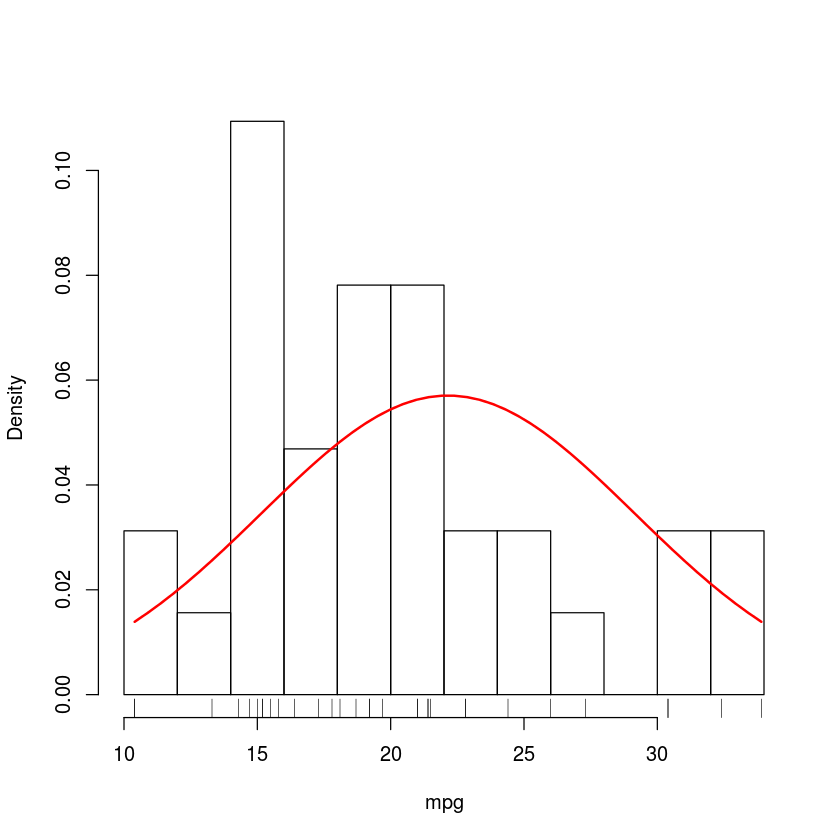
plot(density(mtcars$mpg), main="Density plot")

attach(mtcars)
hist(mpg, breaks=10, probability = TRUE, main="")
rug(mpg)
x <- seq(min(mpg), max(mpg), length.out = 50)
lines(x, dnorm(x, mean=mean(x), sd=sd(x)), col="red", lwd=2)
detach(mtcars)
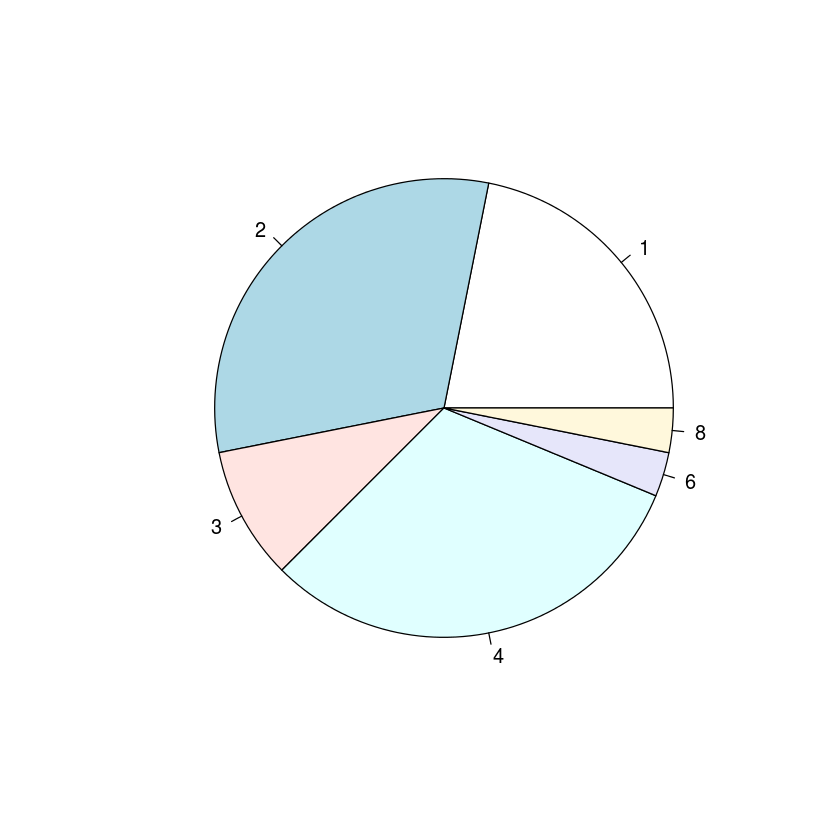
with(mtcars, pie(table(carb)))
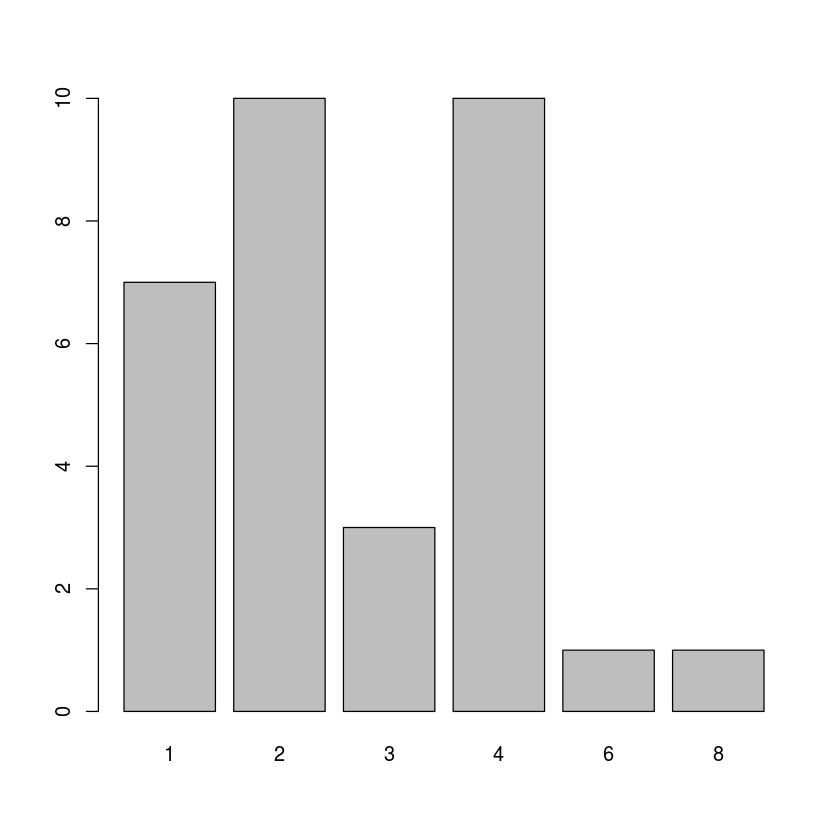
with(mtcars, barplot(table(carb)))
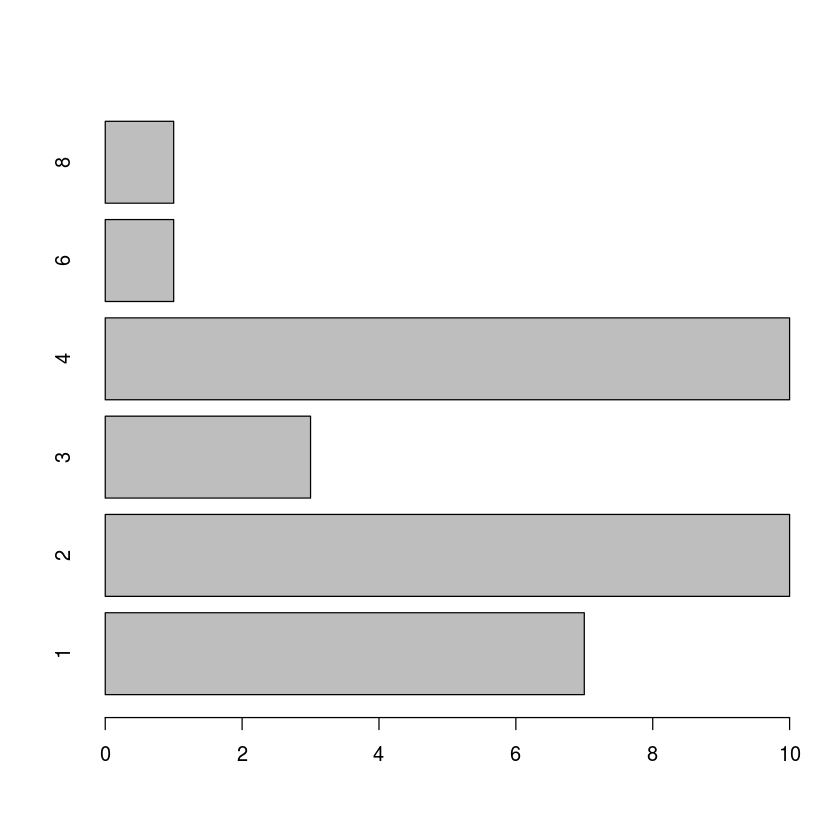
with(mtcars, barplot(table(carb), horiz=TRUE))
attach(mtcars)
(tbl <- table(carb, am))
barplot(tbl, beside=TRUE, legend=rownames(tbl), col=heat.colors(carb))
detach(mtcars)

am
carb 0 1
1 3 4
2 6 4
3 3 0
4 7 3
6 0 1
8 0 1
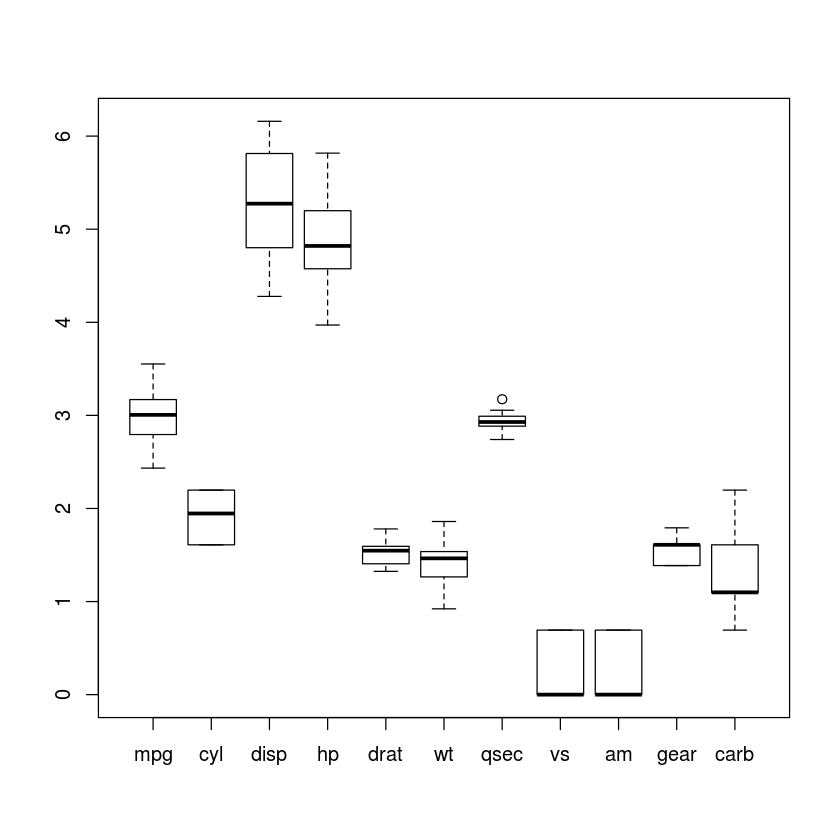
boxplot(log1p(mtcars))
df <- mtcars[order(-mtcars$mpg),]
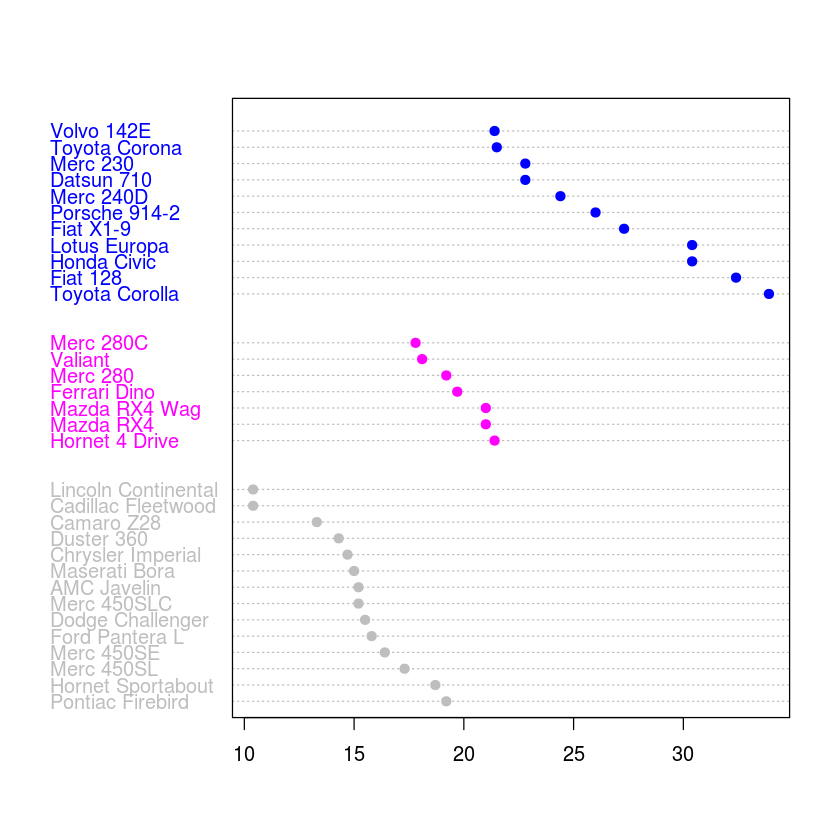
dotchart(df$mpg, labels=row.names(df))

dotchart(df$mpg, labels=row.names(df), groups=df$cyl, color=df$cyl, pch=19)
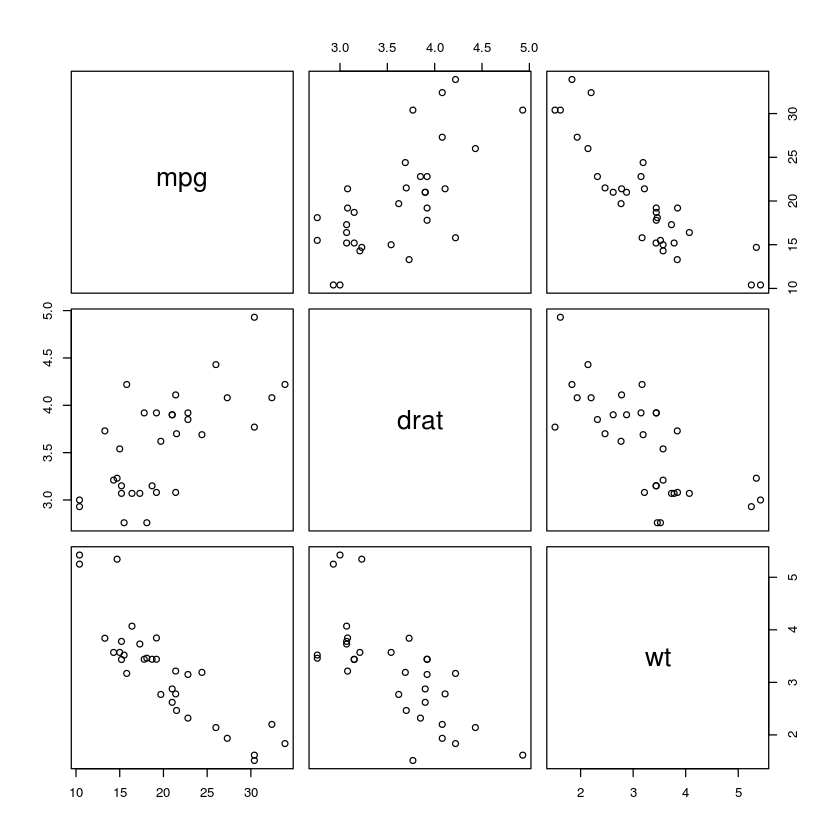
pairs(~mpg + drat + wt, data=mtcars)
Building plots¶
- Local customization of plots
- Glaobl customization of plots
- Adding more stuff to a plot
- points
- lines
- text
- mathematical expressions
- Combining plots
- Saving plots
Combining plots¶
par(mfrow=c(2,2))
plot(rnorm(10))
plot(rnorm(10))
plot(rnorm(10))
plot(rnorm(10))
(m <- matrix(c(1,1,2,3), 2, 2, byrow=TRUE))
| 1 | 1 |
| 2 | 3 |
layout(m)
plot(rnorm(10))
plot(rnorm(10))
plot(rnorm(10))
layout(m, widths=c(1,2), heights=c(2,1))
plot(rnorm(10))
plot(rnorm(10))
plot(rnorm(10))
orig <- par(no.readonly=TRUE)
x <- rnorm(10)
y <- rnorm(10)
par(fig = c(0, 0.8, 0, 0.8))
plot(x, y)
par(fig = c(0, 0.8, 0.6, 1), new=TRUE)
plot(density(x), axes=FALSE, main="", xlab="")
par(fig = c(0.6, 1, 0, 0.8), new=TRUE)
boxplot(y, axes=FALSE)
par(orig)

Plotting Graphcs and Heatmpas¶
Graphs¶
It may be necessary to install first
install.packages("igraphdata", repos = "http://cran.r-project.org")
library(igraph)
library(igraphdata)
data(macaque)
plot(macaque, layout=layout.auto, vertex.shape="circle",
vertex.size=8, edge.arrow.size=0.5, vertex.label=NA)
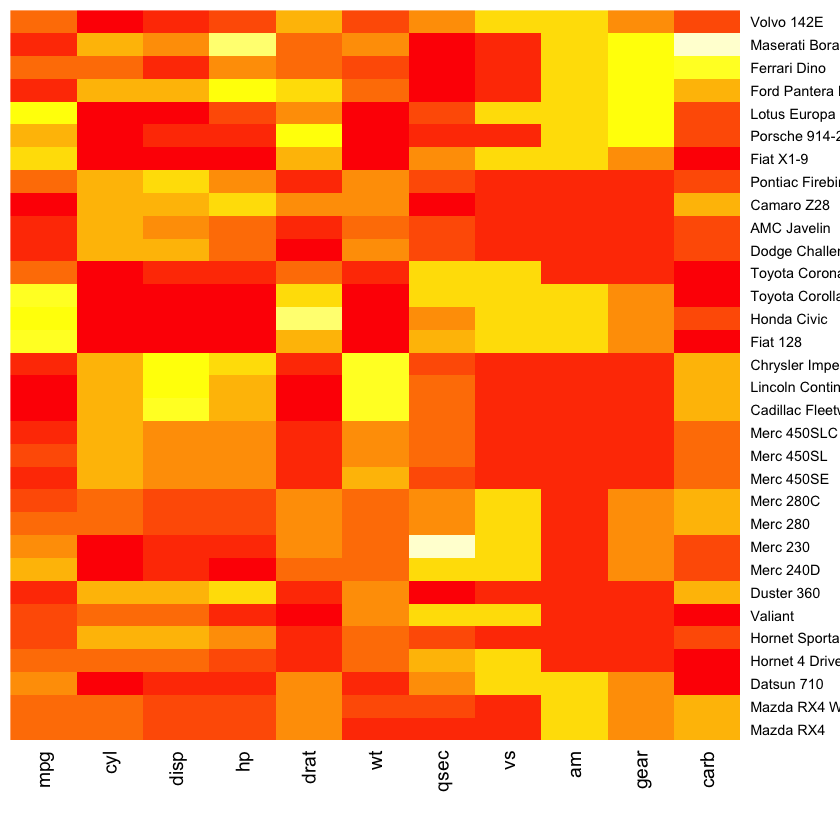
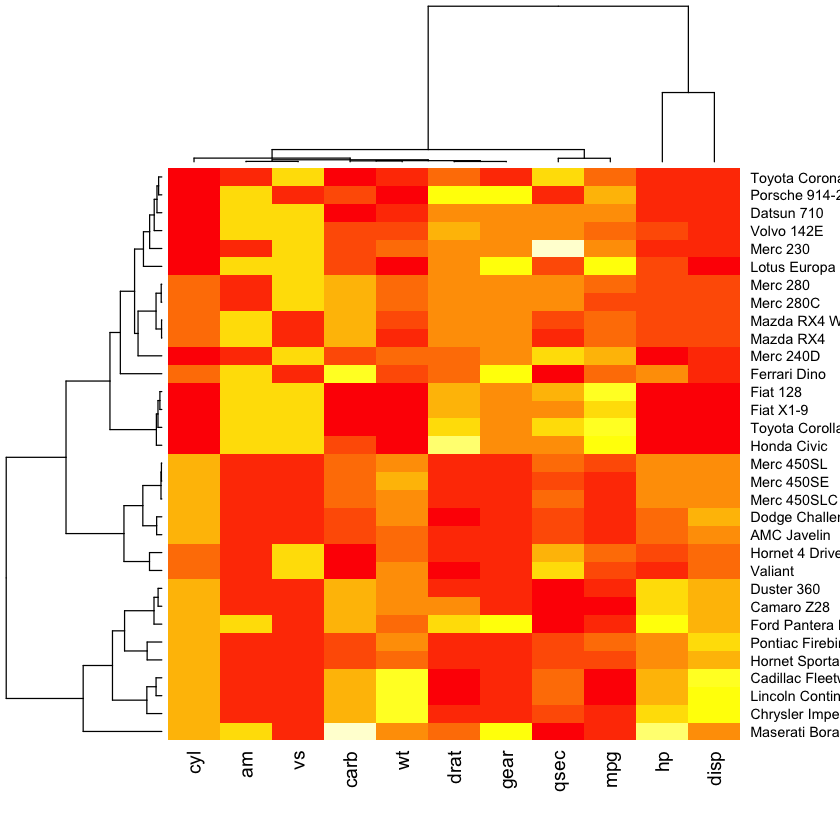
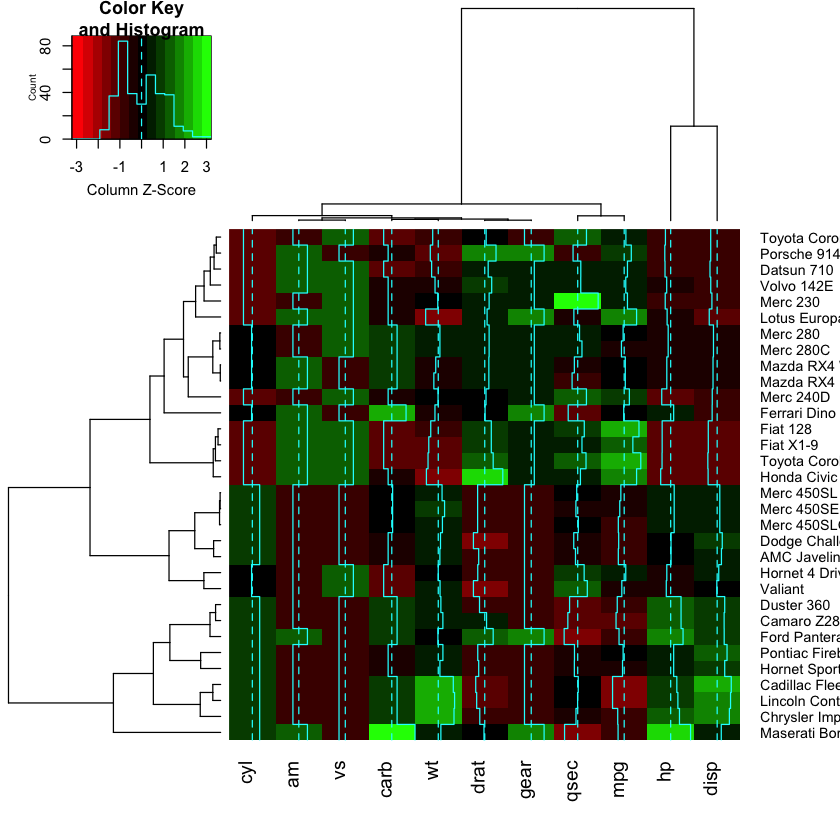
Fancy heatmaps¶
Also see d3heatmaps if you want an interactive heatmap. This does not seem to work within the notebook but will work in RStudio or an R script.
library(pheatmap)
pheatmap(as.matrix(mtcars), scale = "column")

library(gplots)
heatmap.2(as.matrix(mtcars), scale = "column", col=redgreen)
Work!¶
Here we will try to replicate the noise discovery heatmap shown in the statistics class.
suppressMessages(library(genefilter))
Perform noise discovery¶
set.seed(123)
n <- 20 # number of subjects
m <- 20000 # number of genes
alpha <- 0.005 # significance level
# create a matrix of gene expression values with m rows and 2*n columns
M <- matrix(rnorm(2*n*m), m, 2*n)
# give row and column names
rownames(M) <- paste("G", 1:m, sep="")
colnames(M) <- paste("id", 1:(2*n), sep="")
# assign subjects inot equal sized groups
grp <- factor(rep(0:1, c(n, n)))
# calculate p-value using t-test for mean experession value of each gene
pvals <- rowttests(M, grp)$p.value
# extract the genes which meet the specified significance level
hits <- M[pvals < alpha,]
- Use pheatmap to plot a heatmap
- Remove the row names (Google or use R’s built-in help to figure out to do this)
- Use this color palette to map expression values to a red-blakc-green scale
colorRampPalette(c("red3", "black", "green3"))(50)